Modifications of the gut microbiota in chronic inflammatory bowel disease (Crohn’s disease and ulcerative colitis) are both the cause and consequence of these internal disorders. This has been shown a team of French researchers from Inserm, INRA[1], UPMC and AP-HP, who describe these mechanisms and propose new therapeutic approaches. Their work is published in Nature Medicine on 9 May 2016.
(c) Harry Sokol – Inserm
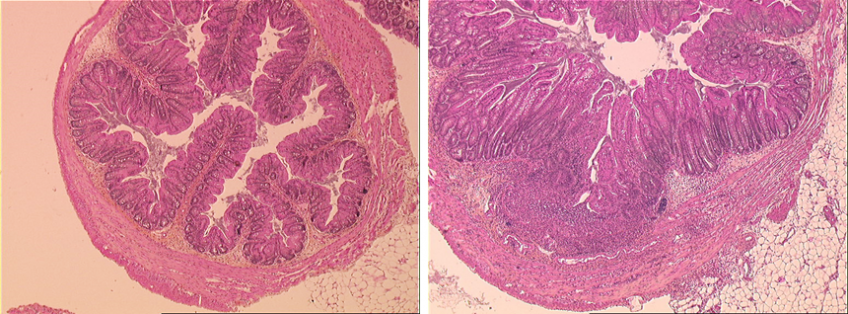
A histology section of axenic, genetically normal mice given the microbiota of genetically normal mice (left) or of Card9−/− mice (right), 12 days after induction of colitis. The colitis is much more severe in mice with the microbiota of Card9−/− mice.
Inflammatory bowel disease (IBD) is characterised by inappropriate inflammation of the digestive tract. It is characterised by inflammatory flare-ups of variable duration and frequency, depending on the patient. This group of diseases usually affects young adults, and its incidence is highest in industrialised countries. Researchers have already discovered susceptibility genes like NOD2, ATG16L1 or CARD9, but also suspect environmental factors and modifications of the gut flora, although when and how these factors are involved is not known.
The CARD9 gene encodes a protein involved in the immune system, and especially in the recognition of microorganisms. “The association between this predisposition gene, the immune system and bacteria deserved to be explored, given that all these factors are involved in IBD,” explains Harry Sokol, leader of this work.
To do this, his team used mice lacking this gene. The researchers found increased sensitivity in the gut of these mice when it was inflamed, with defective healing of the mucosa associated with an interleukin (IL) 22 deficiency and disruptions in the bacterial flora. Observations that did not really surprise them, given that “CARD9 protein expressed by cells of the immune system contributes to the production of IL22, involved in healing and protection of the gut mucosa, and in the recognition of microorganisms,” recalls Harry Sokol.
Except that on transplanting the gut flora from these genetically modified animals to other genetically intact mice but lacking a gut flora, the latter in turn became hypersensitive to gut inflammation. In addition, they also showed a defect in IL22 production. “In other words, the genetic defect in itself is not sufficient to induce the observed malfunctions. Alterations in the composition of the gut flora arising from the absence of CARD9 play a major role in gut hypersensitivity, and the functional defect in the IL22 pathway,” explains Harry Sokol.
The researchers therefore wanted to understand how this altered gut flora could confer these abnormalities on the recipient animal. They then observed that the bacteria present could not, or could only poorly convert tryptophan, an amino acid obtained from food, into an indole derivative that binds to lymphocytes and stimulates IL22 production. An observation that led the researchers to conclude that “Mutation of the CARD9 gene causes a modification of the gut flora mediated by a malfunction of the immune system. The flora loses its ability to produce indole derivatives, helping to exacerbate immunological abnormalities, especially in the IL22 pathway, conducive to inflammation.
These results show how all these mechanisms are interlinked—genetic, immune system and microbiota,” summarises Harry Sokol. “Thus, the abnormalities of the microbiota in IBD are both the cause and consequence of inflammation.”
But most importantly, the researchers showed that these mechanisms were reversible. By giving drugs that can mimic indole derivatives to mice lacking the CARD9 gene, they observed a remission in symptoms, and a return of the IL22 pathway to normal. Exciting results, but it remains to be shown that the same is true of humans. The researchers have already analysed the stools of about a hundred patients with IBD, and have observed a general decrease in the production of indole derivatives by the gut bacteria compared with healthy subjects. By combining this work with a genetic analysis to search for variants of susceptibility genes, they observed that this defect was particularly severe in patients showing a mutation in the CARD9 gene. The idea is now to compensate for this defect in patients. “We can already very easily find patients with a defect in the production of indole derivatives using a simple stool analysis. It may therefore be sufficient to supplement these patients with bacteria producing these derivatives, or directly administer the derivative in question to them.” Work already underway in the laboratory.
[1] “Interactions of commensals and probiotics with the host,” team, MICALIS Institute (Inra-AgroParisTech), Jouy en Josas