A revolution in the field of nanotechnology! An Inserm researcher[1] in collaboration with Harvard University has succeeded in creating 3D shapes of unprecedented sophistication, thanks to the four DNA bases A, T, C and G. In practice, these researchers can create nanoscopic (10-9 m) objects from 30,000 DNA sequences that fold and self-assemble like LEGO® bricks. In time, this will make it possible to manufacture new tools adapted to the size of our cells. These results have been published in Nature.
Nanotechnology represents a rapidly expanding scientific field, particularly when it comes to creating materials with increasingly specific properties. This is the case of carbon nanotubes, for example, which are light but solid and possess very high thermal and electrical conductivity. However, a slightly less well-known field of research exists: that of DNA-based nanotechnology, whose objective is to model living matter to use as a therapeutic tool on a scale compatible with that of a human cell. However, this technology, which was developed in 2012 and is called DNA LEGO® bricks, encountered challenges in programming DNA sequences sufficient to create increasingly complex objects.
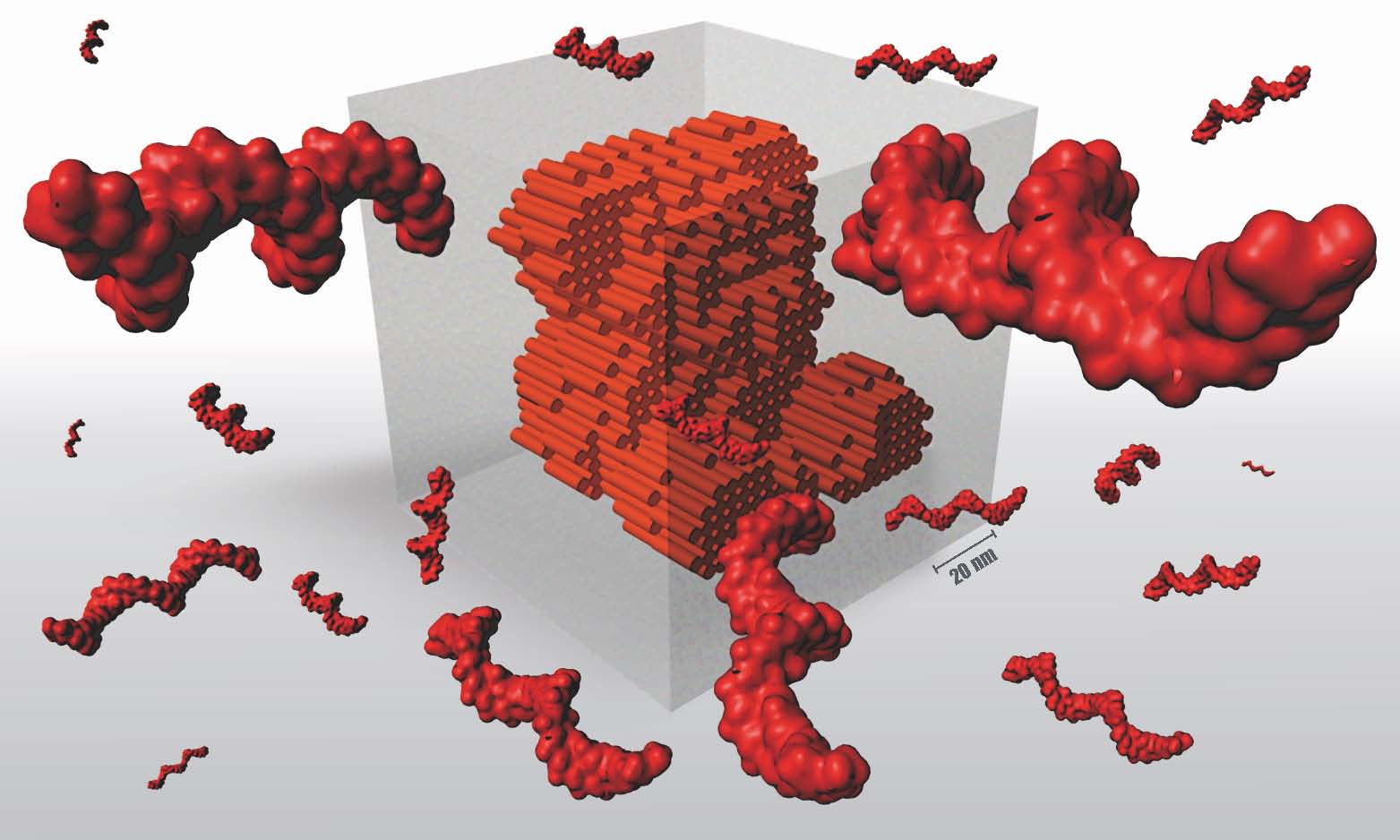
In this paper published in Nature, the researchers have reached a turning point. Their objects, manufactured according to the LEGO® brick method, use one million DNA bases, a size comparable to the genome of a bacterium, whereas until then the objects comprised only one thousand bases.
So, how does it work?
The method uses bricks, like LEGOs®, each comprising 52 DNA bases. One of the properties of DNA is based on the fact that the nucleobases of a DNA strand (A, T, C and G) can interact with those of another strand by always pairing in the same way: A with T, and C with G. Like LEGOs®, these units all have the same general shape but the internal order of the 52 bases determines which bricks will be able to interlock with which and at what level.
Next comes the choice of shape, which is either designed or selected from a database of 3D forms (cube, teddy bear, rabbit, Möbius strip, etc.). Then, each “voxel” [2] of the design is translated into DNA bricks using software developed by the researchers, called Nanobricks. “Nanobricks ‘codes’ the DNA by specifying in advance the order of the 52 bases of each brick that will subsequently be used. This step determines how the 30,000 initial sequences will fit together to produce one final 3D structure,” explains Gaëtan Bellot, Inserm researcher and co-author of this paper.
Once the IT procedures are complete, the 30,000 sequences are synthesized in a laboratory and mixed in a tube. The 30,000 DNA sequences are then completely destructured by means of a denaturing step performed at 80°C. The mixture is then gradually cooled to 25°C at a rate of 0.5°C/hour, which is when the self-assembly takes place. The molecules spontaneously fold and adopt a final shape in accordance with the 3D model selected. In this paper, the researchers produced 13 different objects.
To make objects from 30,000 sequences, they had to increase the diversity of the DNA brick sequences. By exploring various brick sizes, the research teams were able to define an optimal brick size (52 bases) making it possible both to maintain a 3D geometry similar to that of a LEGO brick and increase the diversity of individual bricks to 67 million.
This means that it is possible to obtain increasingly sophisticated objects with a greater diversity of individual bricks. The researchers have succeeded in creating objects with cavities. This level of precision is necessary if we are to design useful and effective tools. “With a key, you can open a car. With a DNA tool you can, for example, build a capsule into which you can place a medicine. And if this object has cavities, you can create a biological chain reaction depending on the products present in each cavity. Taking inspiration from life, this approach will enable the reproduction, on a nanometer scale, of solutions and inventions that occured after millions of years of evolution,” explains Bellot.
This method offers two advantages. The first is that unlike industrial assembly processes, such as car production lines, this technology compresses all the stages into one. Imagine placing a car’s various components in the presence of each other for them to spontaneously self-assemble! The second resides in its rapidity: 30,000 components self-assemble within several hours into an object duplicated a billion times inside the same tube.
Unlike carbon nanotubes, DNA nanotechnologies are biocompatible and can be rapidly eliminated from the human body and the environment. Nevertheless, even if the DNA molecules used are synthetic and as such not biologically active, potential interaction with the DNA present in living organisms cannot be ruled out.
[1] From the Institute for Functional Genomics (Inserm/CNRS/Université de Montpellier)
[2] Voxel, a contraction of the words “volume” and “element”, is a 3D pixel.
