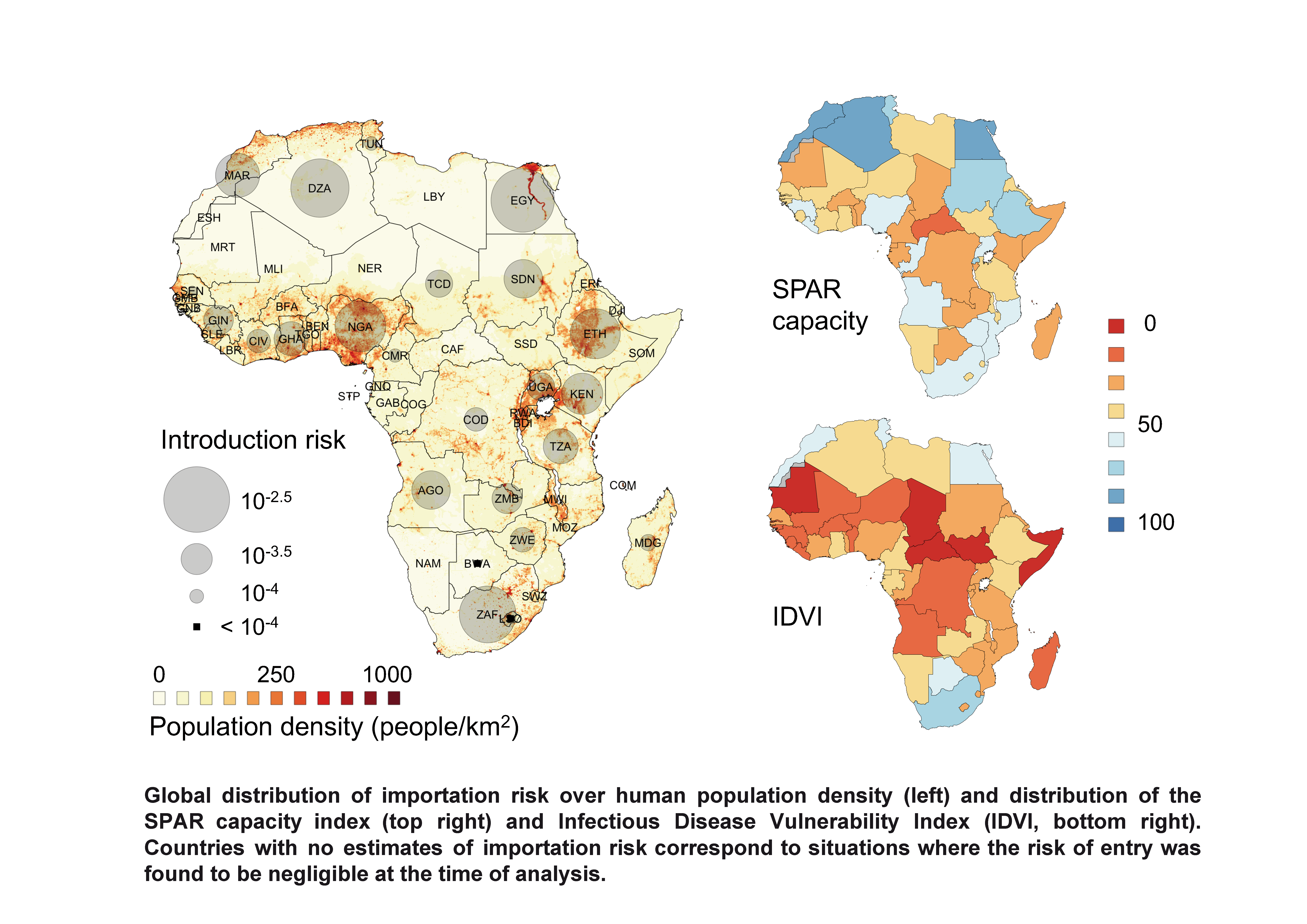
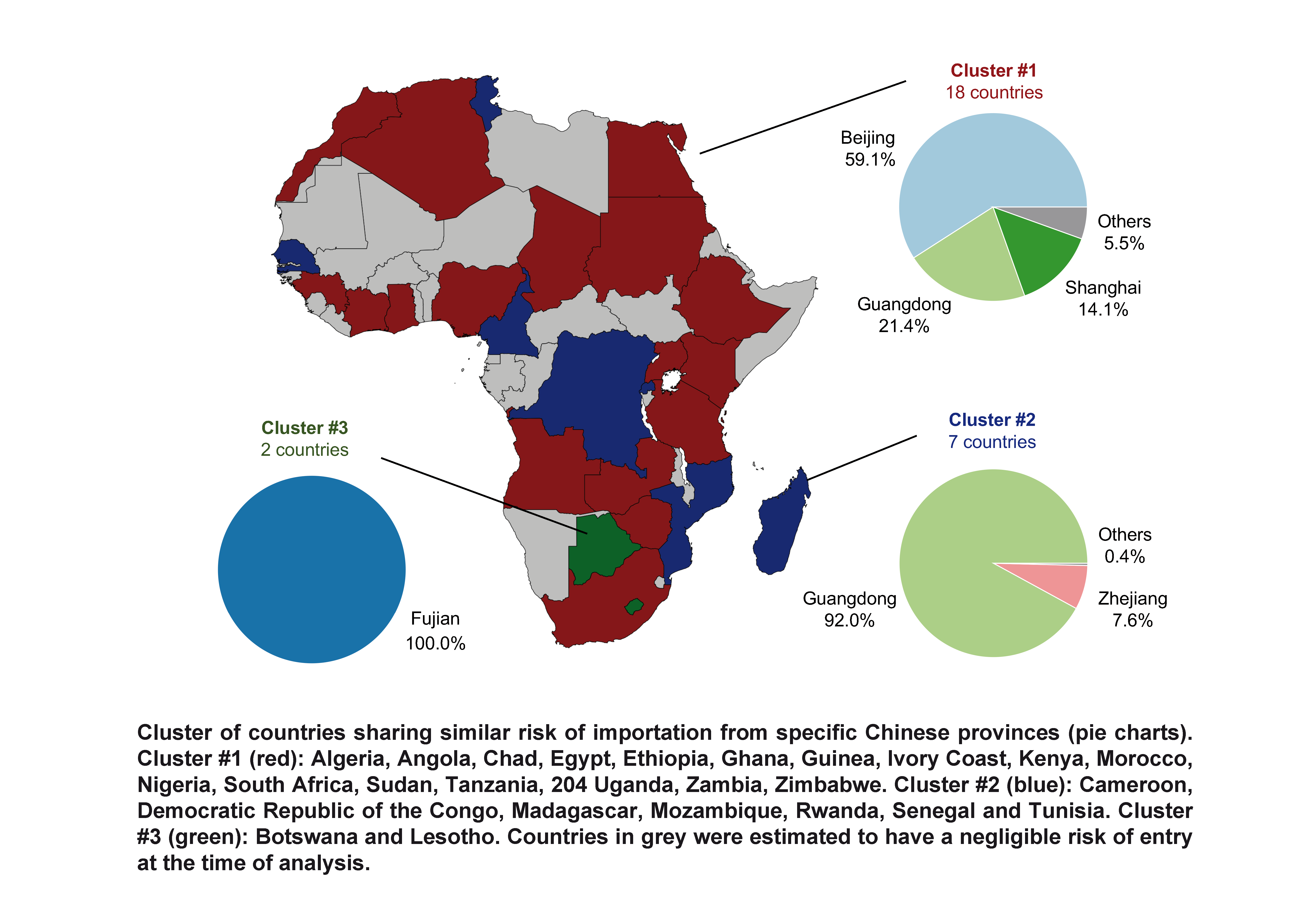
Preparedness and vulnerability of African 1 countries against 2 importations of COVID-19: a modelling study Marius Gilbert1,2,*, Giulia Pullano3,4, Francesco Pinotti3, Eugenio Valdano5, Chiara Poletto3, Pierre-Yves Boëlle3, Eric D’Ortenzio6,7, Yazdan Yazdanpanah6,7, Serge Paul Eholie8.9, Mathias Altmann10,11, Bernardo Gutierrez12, Moritz U.G. Kraemer12,13,14,*, Vittoria Colizza3 1 Spatial Epidemiology Lab (SpELL), Université Libre de Bruxelles, Brussels, Belgium 2 Fonds National de la Recherche Scientifiques, Brussels, Belgium 3 Insem, Sorbonne Université, Institut Pierre Louis d’Epidémiologie et de Santé Publique, IPLESP, Paris, France 4 Sociology and Economics of Networks and Services lab at Orange Experience Design Lab (SENSE/XDLab) Chatillion, Paris, France 5 Center for Biomedical Modeling, The Semel Institute for Neuroscience and Human Behavior, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, United States 6 Université de Paris, IAME, Inserm, F-75018 Paris, France 7 Bichat Claude Bernard Hospital, APHP, Paris, France 8 Service des Maladies Infectieuses et Tropicales, Centre Hospitalier Universitaire de Treichville, Abidjan, Côte d’Ivoire 9 Département de Dermatologie-Infectiologie, Unité de Formation et de Recherche des Sciences Médicales, Université Félix Houphouet-Boigny, Abidjan, Côte d’Ivoire 10 IDLIC – Maladies infectieuses dans les pays à ressources limitées, Inserm U1219, Bordeaux, France 11 Bordeaux Population Health, University of Bordeaux, France 12 Department of Zoology, University of Oxford, Oxford, UK 13 Harvard Medical School, Harvard University, Boston, United States 14 Computational Epidemiology Group, Boston Children’s Hospital, Boston, United States *equal contribution The Lancet : https://doi.org/10.1016/S0140-6736(20)30411-6Preparedness and vulnerability of African 1 countries against 2 importations of COVID-19: a modelling study Marius Gilbert1,2,*, Giulia Pullano3,4, Francesco Pinotti3, Eugenio Valdano5, Chiara Poletto3, Pierre-Yves Boëlle3, Eric D’Ortenzio6,7, Yazdan Yazdanpanah6,7, Serge Paul Eholie8.9, Mathias Altmann10,11, Bernardo Gutierrez12, Moritz U.G. Kraemer12,13,14,*, Vittoria Colizza3 1 Spatial Epidemiology Lab (SpELL), Université Libre de Bruxelles, Brussels, Belgium 2 Fonds National de la Recherche Scientifiques, Brussels, Belgium 3 Insem, Sorbonne Université, Institut Pierre Louis d’Epidémiologie et de Santé Publique, IPLESP, Paris, France 4 Sociology and Economics of Networks and Services lab at Orange Experience Design Lab (SENSE/XDLab) Chatillion, Paris, France 5 Center for Biomedical Modeling, The Semel Institute for Neuroscience and Human Behavior, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, United States 6 Université de Paris, IAME, Inserm, F-75018 Paris, France 7 Bichat Claude Bernard Hospital, APHP, Paris, France 8 Service des Maladies Infectieuses et Tropicales, Centre Hospitalier Universitaire de Treichville, Abidjan, Côte d’Ivoire 9 Département de Dermatologie-Infectiologie, Unité de Formation et de Recherche des Sciences Médicales, Université Félix Houphouet-Boigny, Abidjan, Côte d’Ivoire 10 IDLIC – Maladies infectieuses dans les pays à ressources limitées, Inserm U1219, Bordeaux, France 11 Bordeaux Population Health, University of Bordeaux, France 12 Department of Zoology, University of Oxford, Oxford, UK 13 Harvard Medical School, Harvard University, Boston, United States 14 Computational Epidemiology Group, Boston Children’s Hospital, Boston, United States *equal contribution The Lancet : https://doi.org/10.1016/S0140-6736(20)30411-6