A study carried out by Ali Amara’s team at the combined Inserm/CNRS- Université Paris Diderot “Molecular pathology and virology” unit in the Saint-Louis hospital in Paris, working jointly with the team from the Pasteur Institute in Paris and the team from the Salk Institute in San Diego, has identified two families of receptors that play an important part in the penetration of the Dengue virus into cells. By demonstrating that it is possible to inhibit the viral infection in vitro by blocking the bonding between the virus and these receptors, the researchers have opened the way to a new antiviral strategy. These works were published on line in the review “Cell Host & Microbe” of October 18, 2012.
The Dengue virus circulates in four different forms (four serotypes). It is transmitted to humans by mosquitoes. It is a major public health problem. Two billion people throughout the world are exposed to the risk of infection and 50 million cases of Dengue fever are recorded by the WHO every year. The infection is often asymptomatic, or resembles influenza symptoms, but its most serious forms can lead to fatal haemorrhagic fevers. At present, there is no preventive vaccine or efficient antiviral treatment for these four Dengue serotypes. So it is of vital importance that we develop new therapeutic strategies.
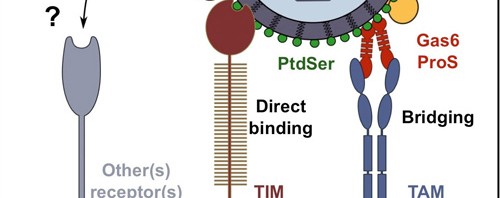
Ali Amara’s team performed genetic screening in order to identify cell receptors used by the virus to penetrate target cells[1]. The researchers have determined the important function played by the TIM receptors (TIM-1, 3, 4) and TAM receptors (AXL and TYRO-3) in the penetration process of the four Dengue serotypes. Mr. Amara’s team has succeeded in demonstrating that the expression of these 2 receptor families makes cells easier to infect. In addition, the researchers observed that interfering RNA or antibodies that target the TIM and TAM molecules considerably reduced the infection of the cells targeted by the Dengue virus. The TIM and TAM molecules belong to two distinct families of transmembrane receptors that interact either directly (TIM) or indirectly (TAM) with phosphatidylserine, an “eat-me” signal that allows the phagocytosis and the elimination of these apoptopic cells. Unexpectedly, the work of the Inserm researchers discovered that phosphatidylserine is abundantly expressed at the surface of virions and that it was essential that the TIM and TAM receptors recognize the phosphatidylserine to allow infection of target cells.
These results have helped to understand the first key stage in the Dengue virus infectious cycle, by discovering a new method of virus entry that works by mimicking the biological functions involved in the elimination of the apoptotic cells. The discovery of these new receptors has also opened the way for new antiviral strategies aimed at blocking bonding of the Dengue virus with the TIM and TAM molecules.
This research has been patent-protected by Inserrm Transfert.
[1] Up till present, only DC-SIGN receptors and L-SIGN receptors were known to play an active role in the penetration of the Dengue virus into target cells