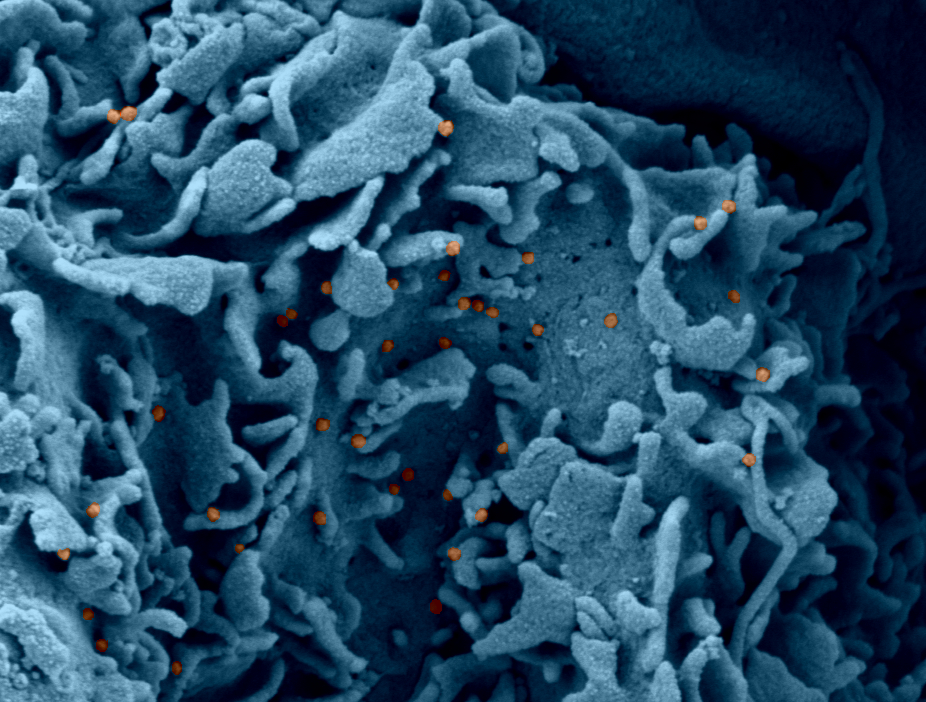
Electron microscopy of a cell infected with SARS-CoV-2 © Philippe Roingeard, Anne Bull-Maurer, Sonia Georgeault, unité Inserm U1259 MAVIVH & Université de Tours, France.
The severe form of Covid-19 is known to be associated with the excessive elevation of many cytokines, a condition termed “cytokine storm”. Therapy with biological agents intended to block these cytokines, for example anti-interleukin-6 or anti-interleukin-1 antibodies, was already tried, albeit with a limited success. However, a study recently published in the Journal of Allergy and Clinical Immunology shows that there are at least two distinct types of cytokine storms induced by SARS-CoV-2 infection that are differentially associated with Covid-19 severity and mortality. As these distinct elevated cytokine profiles show up differently in different patients, this would imply the need for a personalized precision medicine approach to treat Covid-19.
Apart from corticoid therapy, various treatments that were tested to reduce the COVID-19 associated cytokine storm seem to show disappointing results. The effectiveness of reducing mortality by blocking interleukins (for example, IL-6 and IL-1) has so far not been demonstrated. It has also been proposed that early-stage administration of type-I interferon therapy would slow SARS-CoV-2 replication, yet again with only limited reduction in mortality. It is possible that this apparent low effectiveness could be explained by the lack of stratification of patients to receive different treatments adapted to their individual cytokine profile.
© Guy Gorochov
To answer this question, Prof. Guy Gorochov’s team (CIMI Research Center, Sorbonne University / INSERM, Paris, France), in collaboration with Prof. Avidan Neumann’s bioinformatics group (Department of Environmental Medicine, University of Augsburg, Germany), studied the levels of a large number of cytokines in the blood of 115 COVID-19 patients at the day of hospitalization during the first wave of the pandemic. The results of this study were then confirmed by a second validation cohort comprising of 86 patients from the second wave of the SARS-CoV-2 pandemic in Paris1.
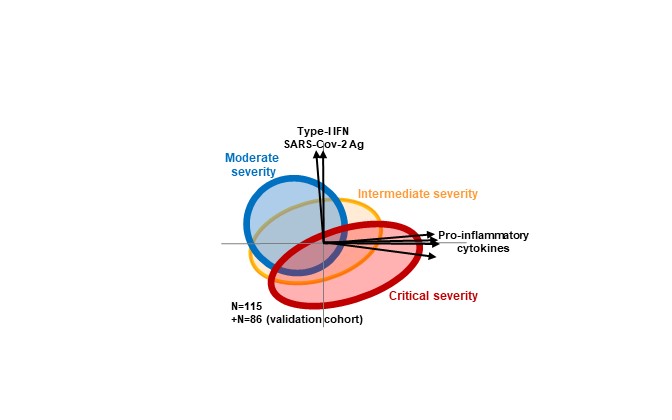
Analysis of the results demonstrated that when looking at each cytokine separately, their blood levels showed great heterogeneity for each individual patient. “By looking at all cytokines together, using a non-supervised bioinformatics method called principle component analysis (PCA), we were able to reduce the large multi-dimensional variation into a two-dimensional perspective of cytokine combinations”, says Prof. Avidan Neumann. “Thus, we identified two distinct cytokine profiles that appear in different levels according to disease severity”.
Moderately severe patients, who initially do not have a severe respiratory illness, develop a response dominated by type-I interferons, in a context of high viral load, and relatively lower levels of pro-inflammatory cytokines. Conversely, patients with critical respiratory severity show elevated levels of pro-inflammatory cytokines (various levels of IL-6, IL-8, IL-10 and TNF-). Unexpectedly, SARS-CoV-2 antigen levels were lower in the critically severe patients, while the interferon anti-viral response is also less prominent in these patients. “These results go against the notion that high COVID-19 severity is always associated with excessive viral replication”, says Prof. Gorochov.
Nevertheless, having a strong type-I interferon response is not always good. An important observation was that the risk of death one month after hospital admission was related in each group to the intensity of the particular cytokine signature typical for that group. In particular, in the moderate severity group mortality was predicted by higher type-I interferon levels, while in the critical severity group mortality was associated with higher pro-inflammatory cytokine levels. Of note, mortality in the critical severity group was best predicted by higher levels of interlukin-10, rather than interlukin-6 or interlukin-1, thus possibly explaining the low effectiveness of therapy blocking the latter cytokines. Moreover, “our results suggest new therapeutic avenues”, says Prof. Gorochov, “as a higher risk of death in the most severe patients requiring ECMO is associated with lower levels of interleukin-17 and interleukin-18, cytokines that are associated with antibacterial response, treatment to increase their levels may improve patients’ survival”.
Overall these results suggest that COVID-19 severity and mortality are associated not with one cytokine storm, but rather with at least two distinct cytokine profiles.
Particularly not only an elevated pro-inflammatory response is dangerous but also, in some patients, an exacerbated anti-viral interferon response is associated with higher risk for mortality.
On the one hand, these results indicate that it would not be necessarily beneficial to administer type-I interferons in patients with already highly elevated levels of these cytokines2. On the other hand, therapy with biological agents should be targeted to block specific pro-inflammatory cytokines that are shown to be elevated in individual patients, rather than a one-for-all therapy. Thus, Prof. Claudia Traidl-Hoffmann, head of the Department of Environmental Medicine at the University of Augsburg, suggests that “this implies a paradigm change in COVID-19 therapy, personalized precision medicine, based on cytokine profiling, should be used to optimize COVID-19 treatment”.
Furthermore, Prof. Neumann adds “generalization of these results to earlier timepoints, currently tested in our on-going Early-Opt project at the Uniklinikum Augsburg, will allow optimization of both public-health and clinical management of COVID-19”.