A team at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC – CNRS/Université de Strasbourg/Inserm) has evidenced, at the atomic scale, the threedimensional structure of the complete human ribosome and the detailed interactions that occur within it. These findings, obtained using a technology that is unique in France, open the way to further exploring some of the adverse effects of antibiotics, and, in the longer term, to the treatment of diseases related to ribosomal dysfunctions and the deregulation of protein synthesis. This work is published in Nature on 22 April 2015.
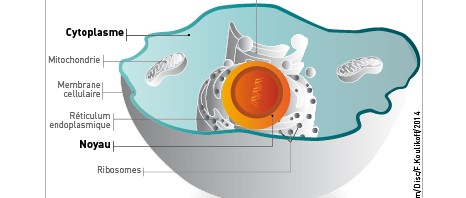
Ribosomes are large complexes of proteins and RNA folded together, which — within the cells of all living beings — act as molecular nano-machines in the expression of genes and the biosynthesis of proteins. While the ribosomal structures of different species were already known in detail at the atomic scale, determining the particularly complex structure of the human ribosome remained a major challenge.
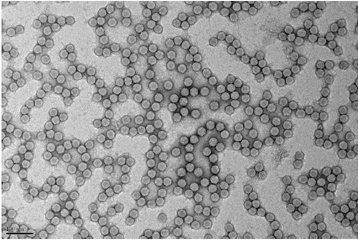
The team led by Bruno Klaholz at IGBMC (CNRS/Université de Strasbourg/Inserm) has now visualized the atomic structure of the complete human ribosome at a resolution greater than 3 angströms (0.3 nanometers). The model obtained represents the 220,000 atoms that make up the two subunits of the ribosome and makes it possible for the first time to explore its detailed arrangement as well as visualize and identify its different amino acids and nucleotides in 3D. The scientists focused in particular on the various binding sites and detailed interactions that occur within this structure. For example, their efforts revealed that after delivering the amino acids they are carrying, transfer RNA continue to interact with the ribosome at a specific site (the tRNA exit site). The team has also shed light on the dynamics of the two ribosomal subunits, which slightly rotate during the protein biosynthesis process, thus heavily remodeling the 3D configuration of the structure at their interface.
These results were achieved using a series of cutting-edge technologies. The samples were highly purified and then frozen before being visualized through cryo-electron microscopy. This method enables scientists to study fixed objects whose orientation does not change and whose structure and biological functions are preserved. A combination of image processing and 3D reconstruction applied to the images obtained by the latest-generation cryo-electron microscope operated by the IGBMC — which is unique in France — made it possible to achieve this rare degree of accuracy.
This detailed knowledge of the structure and dynamics of the complete human ribosome opens the way to further crucial explorations. It is now possible to envisage studying the adverse effects of certain antibiotics designed to fight bacterial ribosomes — and which may target the human ribosome “by mistake”. Listing existing binding sites is the first step towards enhancing the specificity of therapeutic compounds and preventing them from binding to the wrong site. In the longer term, these findings could also help develop treatments for diseases linked to ribosomal dysfunctions and the deregulation of protein synthesis. For example, in the case of cancers, being able to target the ribosomes of diseased cells would make it possible to reduce their protein synthesis rates.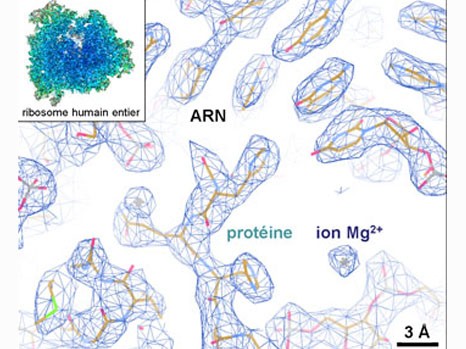
Example of the three-dimensional elements that could be distinguished within the atomic structure of the complete human ribosome (insert). The resolution (of about 1 angström) is able to determine whether or not there are any interactions between the different elements. © H. Khatter, A.G. Myasnikov, S. K. Natchiar & B.P. Klaholz